Jason Kuehner
Associate Professor of Biology - Chair

I am a molecular biologist with a fascination for gene regulation and how cells adapt and respond to stress. Every time we think we have the "dogma" figured out, nature shows us a new trick! I was fortunate to attend a liberal arts college for my undergraduate degree, which provided the freedom to explore academically as well as a mentored faculty-student research experience. My love for learning evolved into a passion for teaching, influenced strongly by a Scientific Teaching Fellowship (HHMI) in graduate school and an IRACDA Postdoctoral Fellowship (NIH/NIGMS). My dream job was to work at a small college in a urban environment, and I found all of that (and more) at Emmanuel! It is a special place where faculty and students can thrive together.
What I Love About Emmanuel:
I love the breadth and depth of learning that is ingrained within the liberal arts and sciences curriculum. I appreciate the personalized connection between faculty and students in the classroom and research laboratory. I relish our dynamic campus community that embraces its rich history and surrounding Boston resources.
- NIH/NIGMS K12 (IRACDA) Postdoctoral Fellowship, Tufts University School of Medicine, Boston, Massachussetts.
- Ph.D., Cellular and Molecular Biology, University of Wisconsin, Madison, Wisconsin.
- B.A., Major in Biochemistry and Molecular Biology (summa cum laude), Minor in Psychology, Cornell College, Mount Vernon, Iowa
- BIOL1105 Introduction to Cellular and Molecular Biology
- BIOL1106 Introduction to Organismic and Evolutionary Biology
- BIOL2123 Genetics
- BIOL2301 Experimental Biology
- BIOL3125 Molecular Biology
- BIOL4160 Biology Senior Seminar
- BIOL4194/95 Research Internship in the Natural Sciences
https://www.ncbi.nlm.nih.gov/myncbi/jason.kuehner.1/bibliography/public/
Recent Research Publications (*indicates Emmanuel undergraduate)
- Pike A, *Pietryski C, Deighan P, Kuehner J, Lau D, Seshan A, March PE. (2024). A simple, robust, broadly applicable insertion mutagenesis method to create random fluorescent protein - target protein fusions. G3: Genes, Genomes, Genetics. Feb 16:jkae036. https://doi.org/10.1093/g3journal/jkae036
- Graber J.H., Hoskinson D., Liu H., Kaczmarek Michaels K., Benson P.S., Maki N.J., Wilson C.L., *McGrath C., Puleo F., Pearson E., Kuehner J.N., Moore C. (2023). Mutations in yeast Pcf11, a conserved protein essential for mRNA 3' end processing and transcription termination, elicit the Environmental Stress Response. Genetics. Nov 15:iyad199. https://doi.org/10.1093/genetics/iyad199
- *Amodeo M.E., *Mitchell S.P.C., *Pavan V., Kuehner J.N. (2023). RNA polymerase II transcription attenuation at the yeast DNA repair gene DEF1 is biologically significant and dependent on the Hrp1 RNA-recognition motif. G3: Genes, Genomes, Genetics. Jan 12;13(1). https://doi.org/10.1093/g3journal/jkac292
- *Whalen, C., *Tuohy, C., *Tallo, T., *Kaufman, J. W., Moore, C., and Kuehner, J.N. (2018). RNA Polymerase II Transcription Attenuation at the Yeast DNA Repair Gene, DEF1, Involves Sen1-Dependent and Polyadenylation Site-Dependent Termination. G3: Genes, Genomes, Genetics, 8: 2043-2058. https://doi.org/10.1534/g3.118.200072
- Kuehner, J.N., *Kaufman, J.W., and Moore, C. (2017). Stimulation of RNA Polymerase II ubiquitination and degradation by yeast mRNA 3’-end processing factors is a conserved DNA damage response in eukaryotes. DNA Repair. 57: 151-160. https://doi.org/10.1016/j.dnarep.2017.07.006
- Graber, J.H., Nazeer, F.I., Yeh, P., Kuehner, J.N., Borikar, S., Hoskinson, D.C., and Moore, C.L. (2013). DNA damage induces targeted, genome-wide variation of poly(A) sites in budding yeast. Genome Research. 23(10): 1690-1703. http://doi.org/10.1101/gr.144964.112
- Gordon, J.M., Shikov, S., Kuehner, J.N., Liriano, M., Lee, E., Stafford, W., Poulsen, M.B., Harrison, C., Moore, C., and Bohm, A. (2011). Reconstitution of CF IA from overexpressed subunits reveals stoichiometry and provides insights into molecular topology. Biochemistry. 50(47): 10203-10214. http://doi.org/10.1021/bi200964p
- Kuehner, J.N., Pearson, E.L., and Moore, C. (2011). Unravelling the means to an end: RNA Polymerase II transcription termination. Nat. Rev. Mol. Cell Biol. 12(5):283-294. http://doi.org/10.1038/nrm3098
- Kuehner, J.N. and Brow, D.A. (2008). Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell. 31(2): 201-211. [Selected as cover article; Highlighted in Preview, Dichtl, B. (2008). Mol. Cell.] http://doi.org/10.1016/j.molcel.2008.05.018
- Steinmetz, E.J., Warren C.L., Kuehner, J.N., Panbehi, B., Ansari, A.Z., and Brow, D.A. (2006). Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell. 24(5): 735-746. [Selected as cover article]. http://doi.org/10.1016/j.molcel.2006.10.023
- Kuehner, J.N. and Brow, D.A. (2006). Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. J. Biol. Chem. 281(20): 14119-14128. [Highlighted in News and Views, Hampsey, M., (2006). Nat. Struct. Mol. Biol.] http://doi.org/10.1074/jbc.M601937200
Teaching Publications
- Cloud-Hansen, K.A.#, Kuehner, J.N.#, Tong L., Miller, S., and Handelsman, J. (2008). Money, sex, and drugs: a case study to teach the genetics of antibiotic resistance. CBE Life Sci. Educ. 7(3): 302-309. http://doi.org/10.1187/cbe.07-12-0099
#authors contributed equally to this work
Recent Research Presentations (*indicates Emmanuel undergraduate)
International + National Conferences
- *Talluto, J., *Richa, J., *Lapine, M., and Kuehner, J.N. (2024). Attenuation of RNA Polymerase II transcription at multiple 5’-end RNA sequences requires the yeast 3’-end processing factor Hrp1 and its RNA Recognition Motif. RNA Society Meeting. Edinburgh, Scotland. (Poster)
- *Roche, M., *Edouard, S., *Talluto, J., *Pavan, V., and Kuehner. J.N. (2023). RNA Polymerase II transcription attenuation at multiple genes in yeast depends on the mRNA 3’-end processing factor Hrp1 and its RNA recognition motif. America Society for Biochemistry and Molecular Biology – Annual Meeting. Seattle, WA. (Poster) https://doi.org/10.1016/j.jbc.2023.103408
- *Amodeo M.E., *Mitchell S.P.C., *Pavan V., and Kuehner J.N. (2022). RNA polymerase II transcription attenuation at the yeast DNA repair gene DEF1 is biologically significant and dependent on the Hrp1 RNA-recognition motif. CNRS Conference: Gene Transcription in Yeast. Sant Feliu de Guizols, Spain. (Platform talk)
- *Mitchell, S. and Kuehner, J.N. (2019). Investigating the biological significance of RNA Polymerase II attenuation at the yeast DNA repair gene, DEF1. International Conference on Yeast Genetics and Molecular Biology. Götenburg, Sweden. (Poster)
- *Whalen, C., *Tuohy, C., *Tallo, T.W., *Kaufman, J.W., Moore, C., and Kuehner, J.N. (2018). RNA Polymerase II transcription attenuation at the yeast DNA repair gene, DEF1, involves Sen1-dependent and polyadenylation site-dependent termination. EMBO Conference: Gene Transcription in Yeast. Sant Feliu de Guizols, Spain. (Session Chair and Poster)
- Kuehner, J.N., *Kaufman, J., and Moore, C.L. (2016). Stimulation of RNA Polymerase II ubiquitination by yeast RNA 3’ processing factors is a conserved DNA damage response in eukaryotes. Yeast Genetics and Molecular Biology Conference, Orlando, FL. (Poster)
- Kuehner, J.N., *Tallo, T.W., *Kelly, K.E., and Moore, C.L. (2015). Stimulation of RNA Polymerase II ubiquitination by yeast RNA 3’ processing factors is a conserved DNA damage response in eukaryotes. Mechanisms of Eukaryotic Transcription Conference, Cold Spring Harbor, NY. (Poster)
- Kuehner, J.N., *Duffy H., and Moore, C.L. (2014). Stimulation of RNA Polymerase II ubiquitination by yeast RNA 3’ processing factors is a conserved DNA damage response in eukaryotes. Yeast Genetics and Molecular Biology Conference, Seattle, WA. (Poster)
- Kuehner, J.N., Nazeer, F.I., and Moore, C.L. (2011). Coupling of RNA 3’-end processing with the DNA damage response. Canadian RiboClub Conference. Orford, Quebec. (Platform talk and Poster)
- Kuehner, J.N. and Moore, C.L. (2010). Mutations in the yeast nuclear mRNA cap-binding protein Cbp80 suppress a Pcf11 mutant defective for binding RNA Polymerase II. Gordon Conference: Post-transcriptional Gene Regulation. Newport, RI. (Poster)
- Kuehner, J.N. (2009). EMBO Conference: Messenger RNA 3’-ends and Gene Expression. Oxford, England. (Participant)
- Kuehner, J.N. and Brow, D.A. (2008). Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. EMBO Conference: Gene Transcription in Yeast. Sant Feliu de Guizols, Spain. (Platform talk)
- Kuehner, J.N. and Brow, D.A. (2007). Sen1-dependent attenuation of yeast RNA polymerase II transcription. RNA Society Conference, Madison, WI. (Platform talk)
- Kuehner, J.N., Steinmetz, E.J., and Brow, D.A. (2007). Sen1-dependent attenuation of yeast RNA polymerase II transcription. Cold Spring Harbor Conference: Mechanisms of Eukaryotic Transcription. Cold Spring Harbor, NY. (Poster)
- Kuehner, J.N. and Brow, D.A. Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. Yeast Genetics and Molecular Biology Conference, Princeton, NJ. (2006). (Poster)
Regional Conferences
- *Richa, J. and Kuehner, J.N. (2024). Taking the bait: Capturing the interplay of transcription termination protein Hrp1 with multiple RNA targets. Eastern New England Biological Conference, Merrimack College, North Andover, MA. (Poster)
- *Talluto, J. and Kuehner, J.N. (2024). To bind or not to bind: Characterizing RNA-Protein interactions during transcription termination. Eastern New England Biological Conference, Merrimack College, North Andover, MA. (Poster)
- *Roche, M. and Kuehner, J.N. (2023). Investigating the Hrp1 interaction with RNA binding sites via mutations in proteins and RNA. Eastern New England Biological Conference, Simmons University, Boston, MA. (Poster)
- *Edouard, S. and Kuehner, J.N. (2023). Identifying sequence and protein requirements for premature transcription termination (attenuation) of RNA Polymerase II at the yeast gene SNG1. Eastern New England Biological Conference, Simmons University, Boston, MA. (Poster)
- *Amodeo, M. and Kuehner, J.N. (2021). Cutting the cellular brakes: Characterizing the role of Hrp1 in pre-mature transcription termination. TriBeta NE-1 District Convention. Virtual. (Winner of Frank G. Brooks Award for Best Platform Talk).
- *Amodeo, M. and Kuehner, J.N. (2019). Cutting the cellular brakes: Investigating removal of transcription termination factor Hrp1. Emmanuel College Research Symposium. (Winner of Hakim Scholarship for “Best Student Research Poster, School of Science and Health”).
- *Welsh, C. and Kuehner, J.N. (2019). I will find you and I will degrade you: Analysis of transcription termination factor depletion. Eastern New England Biological Conference, Emmanuel College, Boston, MA. (Poster)
- *McGovern, A. and Kuehner, J.N. (2019). Is all stress the same?: Characterizing transcriptional termination responses to salt, stress, and heat. Eastern New England Biological Conference, Emmanuel College, Boston, MA. (Poster)
- *McGrath, C. and Kuehner, J.N. (2018). Caught at a red light: Regulation of DNA transcription by premature termination. Eastern New England Biological Conference, Colby-Saywer College, New London, NH. (Poster)
- *Kaufman, J. and Kuehner, J.N. (2018). Who flipped the switch? – Investigating regulation of the yeast DNA repair gene, DEF1, by premature transcription termination. Eastern New England Biological Conference, Colby-Sawyer College, New London, NH. (Winner of 2nd place “Best Poster Award”)
- *Whalen, C., Moore, C., and Kuehner, J.N. (2017). You are terminated: Premature transcription stoppage regulates yeast gene expression following DNA damage. Eastern New England Biological Conference, Suffolk University, Boston, MA. (Winner of “Top 3 Best Poster Award”)
- *Tuohy, C., *Tallo, T. and Kuehner, J.N. (2016). Identification of cis-acting elements required for transcriptional attenuation of the yeast DNA repair gene DEF1. Eastern New England Biological Conference, Quinnipiac University, Hamden, CT. (Poster)
- *Tallo, T., *Tuohy, C. and Kuehner, J.N. (2016). Identification of trans-acting factors required for transcriptional attenuation of the yeast DNA repair gene DEF1. Eastern New England Biological Conference, Quinnipiac University, Hamden, CT. (Poster
- *Brioso, S.M.T., Moore, C.L., and Kuehner, J.N. (2015). Investigating the role of RNA 3’-end processing factors in the yeast cell wall integrity pathway. Eastern New England Biological Conference, Boston, MA. (Poster)
- *Kelly, K.E., *Tallo, T.W., Moore, C.L., and Kuehner, J.N. (2015). The secret life of RNA processing factors: Investigating their role in repairing UV-damaged DNA. Eastern New England Biological Conference, Boston, MA. (Poster)
Selected Education Conferences and Workshops
- *Roche, M., *Edouard, E., *Talluto, J., and Kuehner, J.N. (2023). Experimental Biology CURE Lab: Hunting for Transcriptional Attenuators that Regulate Eukaryotic Gene Expression. American Association for Biochemistry and Molecular Biology Conference – Transforming Undergraduate Education in the Molecular Life Sciences. Suffolk University, Boston, MA. (Poster)
- BREWMOR (Bridging Research and Education With Model ORganisms) Workshop. (2021). “Incorporating hands-on research, including model organisms, into the undergraduate classroom.” Virtual Meeting. (Participant)
- Colleges of the Fenway Teaching and Learning Conference. (2019). Teaching in a Radicalized Environment. Massachusetts College of Art and Design, Boston, MA. (Participant)
- Jarvinen, M., Johnston, L., Kuehner, J.N., Leighton, C.M., Colleges of the Fenway Teaching and Learning Conference. (2018). Teaching, Research, and Scholarship at its best within the COF. Emmanuel College. Boston, MA. (Platform presentation)
- Colleges of the Fenway Teaching and Learning Conference. (2017). Self-regulated learning. Emmanuel College. Boston, MA. (Participant)
- Kuehner, J.N. and Dementieva, Y. (2017). The search for significance: Integrating statistics and genetics in the modern sequencing era. Genome Consortium for Active Teaching – Next Generation Sequencing Group (GCAT-SEEK). Hampton. (Selected participants)
- Colleges of the Fenway Teaching and Learning Conference. (2016). Understanding and Teaching Today’s Students. Massachusetts College of Art and Design. Boston, MA. (Participant)
- Colleges of the Fenway Teaching and Learning Conference. (2015). Critical Conversations on Campus. Simmons College, Boston. (Participant)
- Colleges of the Fenway Teaching and Learning Conference. (2014). Improving Student Learning Through Metacognition. Emmanuel College, Boston, MA. (Participant)
- Colleges of the Fenway Teaching and Learning Conference. (2013). 21st Century Learning – Implications for College Teaching. Simmons College, Boston, MA. (Participant)
- Colleges of the Fenway Teaching and Learning Conference. (2012). What the Best Colleges Teachers Do - And How You Can Be Among Them. Wheelock College, Brookline, MA. (Participant)
- Institutional Research and Academic Career Development Award (IRACDA) Conference, Houston, TX (2011). Coupling of RNA 3’-end processing with the DNA damage response. (Poster)
- Institutional Research and Academic Career Development Award (IRACDA) Conference. (2010). Boston, MA. (Participant)
- Institutional Research and Academic Career Development Award (IRACDA) Conference. (2009). Mutations in the yeast nuclear mRNA cap-binding protein Cbp80 suppress a Pcf11 mutant defective for binding RNA Polymerase II. San Francisco, CA. (Poster)
PI External Funding
- National Science Foundation Research in Undergraduate Institutions (NSF-RUI) Grant, 2022-2025 ($372,026)
- LI-COR – Science Undergraduate Research Grant (SURG), 2017 ($3,238)
- National Institutes of Health – National Research Mentoring Network Fellowship (NIH-NRMN), 2016 ($9,875)
- National Science Foundation – Research Opportunity Award (NSF-ROA), 2015 ($37,432)
Student External Funding
- Tri-Beta Biological Honor Society Grant (6 students; Total funding: $4,179)
- Sigma Xi Scientific Research Honor Society ($859)
PI Awards + Recognition
- American Society for Biochemistry and Molecular Biology Travel Award (2023)
- Emmanuel College Faculty Excellent in Teaching Award (2018)
- Office of Student Activities and Multicultural Programs
- Faculty Advisor of the Year Nomination (2015, 2017, 2019)
- “Impact” Award (2016)
- Academic Technology and Innovation Group “Best ECLearn Course Website” (2016)
- NIH/NIGMS K12 Postdoctoral Fellowship (2008-2011)
- EMBO Workshop Travel Award (2008)
- HHMI Teaching Fellow in Classroom Teaching and Mentoring (2006)
- Genetics Society of America Travel Award (2006)
- Cornell College Outstanding Senior Award Biochemistry & Molecular Biology (2002)
- Phi Bea Kappa Honor Society (2001)
(General Description) Life depends on information stored in DNA, which is expressed into RNA and proteins that perform cellular functions. An initial step of gene expression is transcription, where a molecular machine called RNA polymerase reads DNA to synthesize RNA. As RNA polymerase moves along DNA, it can be interrupted by regulatory stop signals that terminate transcription prematurely. Downregulation of gene expression by early transcription stoppage occurs widely in cells ranging from bacteria to human, but the underlying mechanism and selectivity remains unclear. This research project will investigate premature transcription termination in the yeast S. cerevisiae, a tractable model for studying many conserved biological processes. Undergraduate student researchers will be trained to use classical genetics and modern bioinformatic tools to dissect transcription stop signals and discover mutants that alter recognition. New gene targets will be identified, with a broader goal of identifying shared regulatory features. Student trainees will have opportunities to present their work at national research conferences and coauthor publications. This research will also be incorporated into a core undergraduate biology laboratory course, mobilizing 50 additional students to validate gene targets. Course resources will be shared broadly so additional educational communities may contribute and benefit.
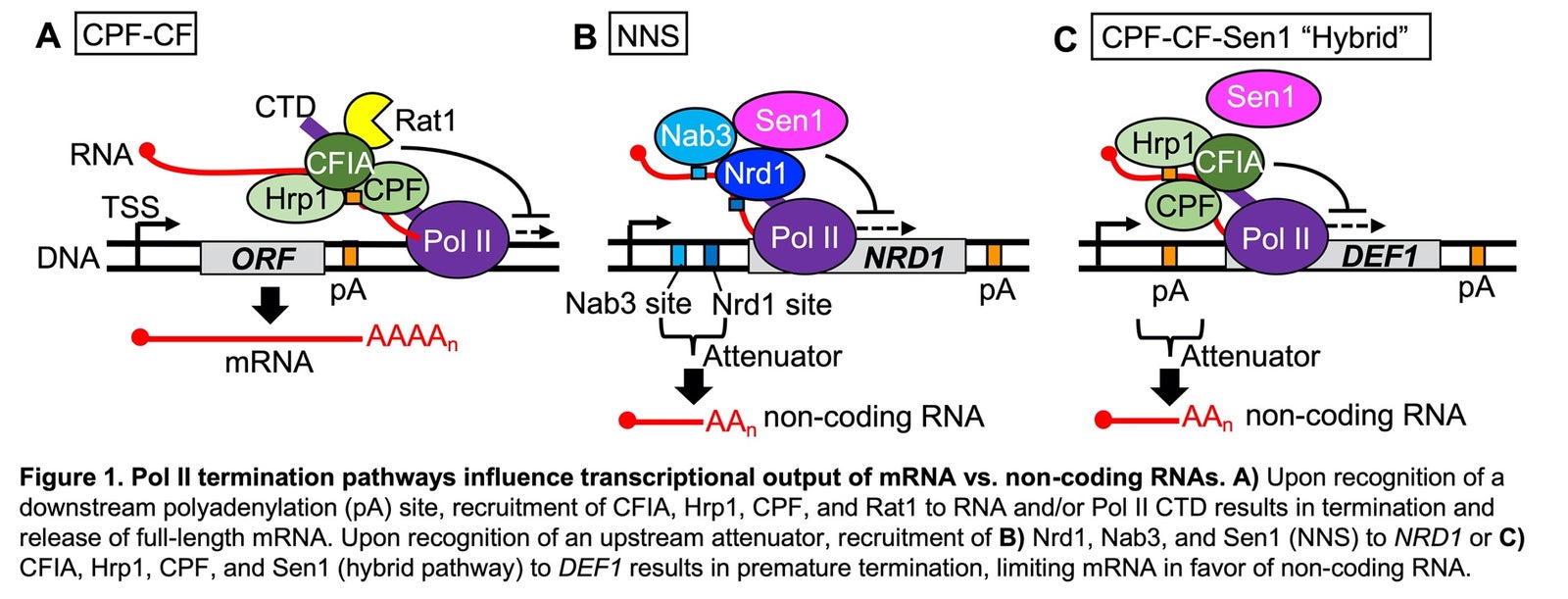
(Specialist Description) Gene regulation is integral to biological control, and transcription is one of the earliest and most powerful steps to modulate gene expression. During productive transcription of mRNA genes in eukaryotes, RNA Polymerase II (Pol II) initiates at a promoter and elongates RNA through the gene open reading frame (ORF). Pol II transcription termination is coupled with mRNA 3’-end processing and polyadenylation, which has been best studied in the model eukaryote yeast (Fig. 1A). Gene downregulation can occur via premature transcription termination of RNA polymerase (i.e. attenuation), which limits synthesis of full-length mRNA and thereby restricts protein production (Fig. 1B). As a testament to its biological utility, attenuation is one of the most ancient and widespread forms of gene regulation, spanning all three domains of life and viruses. Once thought to be rare, attenuation of Pol II transcription in eukaryotes appears even more prevalent than in bacteria, occurring at 10-15% of mRNA genes in yeast and higher eukaryotes. However, the mechanism and selectivity of Pol II attenuation remains unclear. Our lab recently helped characterize a hybrid attenuation pathway involving Hrp1, an RNA-binding protein in the 3’-end cleavage factor (CF) complex, and the Sen1 helicase (Fig. 1C). The hybrid pathway appears to be an alternative to the canonical attenuation pathway in yeast, which relies on the RNA-binding proteins Nrd1/Nab3 and Sen1 (NNS). We hypothesize that Hrp1-dependent hybrid termination contributes broadly to yeast Pol II attenuation.
Research Significance
Our study of Pol II attenuation is significant because it will increase understanding of how RNA-based gene regulation evolves over time. Our work will inform analysis of the Hrp1 ortholog HNRNPDL, a human protein that likewise binds AU-rich RNA and regulates transcription. In addition, naturally-occurring and artificially-engineered attenuators may be harnessed for dynamic gene control in biotechnology applications, including yeast expression of industrial enzymes.
Kuehner Lab Approach
Using a combination of molecular biology (e.g. PCR, Gibson cloning), genetics (e.g. mutagenesis, CRISPR), biochemistry (e.g. lacZ reporter assay), and bioinformatics (genome browser analysis), we aim to generate a comprehensive profile of Pol II attenuation determinants (e.g. RNA-binding elements and protein recognition factors) and identify a myriad of new attenuators for further study. We ultimately hope to uncover novel attenuation mechanisms in the yeast model system that may be shared across species. In addition we aim to train future STEM professionals, and Kuehner lab alumni have successfully pursued careers in research (academia, industry), health care (PA, NP, RN), and government.